In mathematics, linear interpolation is a method of curve fitting using linear polynomials to construct new data points within the range of a discrete set of known data points.[1]
In [1]:
%matplotlib inline
%watermark -dtvmp numpy,matplotlib,pandas,cython
import matplotlib.pylab as plt
import lerp
from lerp import *
2017-10-12 22:57:48
CPython 3.6.2
IPython 6.1.0
numpy 1.13.1
matplotlib 2.0.2
pandas 0.20.3
cython 0.26.1
compiler : GCC 4.2.1 Compatible Clang 4.0.1 (tags/RELEASE_401/final)
system : Darwin
release : 15.6.0
machine : x86_64
processor : i386
CPU cores : 2
interpreter: 64bit
In [2]:
lerp.options.display.max_rows = 15
Usage¶
BreakPoints¶
In [3]:
A = BreakPoints(d=[1.040, 1.051, 1.057, 1.063, 1.064, 1.067, 1.068, 1.068, 1.068, 1.066, 1.064,
1.060, 1.056, 1.050, 1.042, 1.032], label="Ballistic coefficient", unit="G1")
Display in the notebook, integer above the value are helps for indexing purpose
In [4]:
A
Out[4]:
BreakPoints: Ballistic coefficient [G1]
| 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | ... | 15 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1.04 | 1.051 | 1.057 | 1.063 | 1.064 | 1.067 | 1.068 | 1.068 | 1.068 | 1.066 | 1.064 | 1.06 | 1.056 | 1.05 | ... | 1.032 |
In [5]:
A[6]
Out[5]:
1.0680000000000001
Get a plot with the plot() method
In [6]:
A.plot()
Out[6]:
[<matplotlib.lines.Line2D at 0x181b8a0400>]
Not that fancy… you can set the ggplot style
In [7]:
plt.style.use('ggplot')
A.plot()
Out[7]:
[<matplotlib.lines.Line2D at 0x181ba223c8>]
If you don’t like ggplot, choose one of the seaborn styles
In [8]:
import matplotlib.gridspec as gridspec
import matplotlib as mpl
styles = [_s for _s in plt.style.available if 'seaborn' in _s]
n = np.ceil(np.sqrt(len(styles))).astype(np.int)
gs = gridspec.GridSpec(n, n)
plt.figure(figsize=(24,18))
for i, s in enumerate(styles):
mpl.rcParams.update(mpl.rcParamsDefault)
plt.style.use(s)
plt.subplot(gs[i])
A.plot()
(A / 1.01).plot()
(A / 1.02).plot()
plt.title(s)
plt.tight_layout()
mesh2d¶
From Ballistic coefficient article in Wikipedia.
Doppler radar measurement results for a lathe turned monolithic solid .50 BMG very-low-drag bullet (Lost River J40 13.0 millimetres (0.510 in), 50.1 grams (773 gr) monolithic solid bullet / twist rate 1:380 millimetres (15 in)) look like this:
In [9]:
BC = mesh2d(x=np.arange(500,2100,100), x_label="Range", x_unit="m",
d=[1.040, 1.051, 1.057, 1.063, 1.064, 1.067, 1.068, 1.068, 1.068, 1.066, 1.064,
1.060, 1.056, 1.050, 1.042, 1.032], label="Ballistic coefficient", unit="G1")
Display in the jupyter notebooks / ipython
In [10]:
BC
Out[10]:
mesh2d
| 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | ... | 15 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Range [m] | 500 | 600 | 700 | 800 | 900 | 1000 | 1100 | 1200 | 1300 | 1400 | 1500 | 1600 | 1700 | 1800 | ... | 2000 |
| Ballistic coefficient [G1] | 1.04 | 1.051 | 1.057 | 1.063 | 1.064 | 1.067 | 1.068 | 1.068 | 1.068 | 1.066 | 1.064 | 1.06 | 1.056 | 1.05 | ... | 1.032 |
Interpolation
In [11]:
BC(501)
Out[11]:
1.04011
In [12]:
BC([501, 609, 2500])
Out[12]:
array([ 1.04011, 1.05154, 0.982 ])
Default : values are extrapolated
In [13]:
BC.interpolate([501, 609, 2500])
Out[13]:
array([ 1.04011, 1.05154, 1.032 ])
Interpolation is performed and boundaries values are kept
In [14]:
BC.options
Out[14]:
{'extrapolate': True, 'step': False}
In [15]:
BC.options['extrapolate']= False
This can be controled though the dict key extrapolate in options or interpolate method.
In [16]:
BC([501, 609, 2500])
Out[16]:
array([ 1.04011, 1.05154, 1.032 ])
In [17]:
BC.max()
Out[17]:
1.0680000000000001
In [18]:
BC.max(argwhere=True)
Out[18]:
(1100, 1.0680000000000001)
Plot as steps
I like the color from vega
In [19]:
from cycler import cycler
category20 = cycler('color', ['#1f77b4', '#aec7e8', '#ff7f0e',
'#ffbb78', '#2ca02c', '#98df8a',
'#d62728', '#ff9896', '#9467bd',
'#c5b0d5', '#8c564b', '#c49c94',
'#e377c2', '#f7b6d2', '#7f7f7f',
'#c7c7c7', '#bcbd22', '#dbdb8d',
'#17becf', '#9edae5'])
plt.style.use('seaborn-darkgrid')
plt.rc('axes', prop_cycle=category20)
In [20]:
plt.figure(figsize=(10,10))
BC.plot(label="Interpolation between points")
BC.steps.plot(label="Steps")
BC.steps.plot(where="pre", label="Steps, pre")
plt.graphpaper(dx=200, dy=0.005)
plt.legend(bbox_to_anchor=(1.05, 0.5), loc='center left', facecolor="white", frameon=False)
plt.tight_layout()
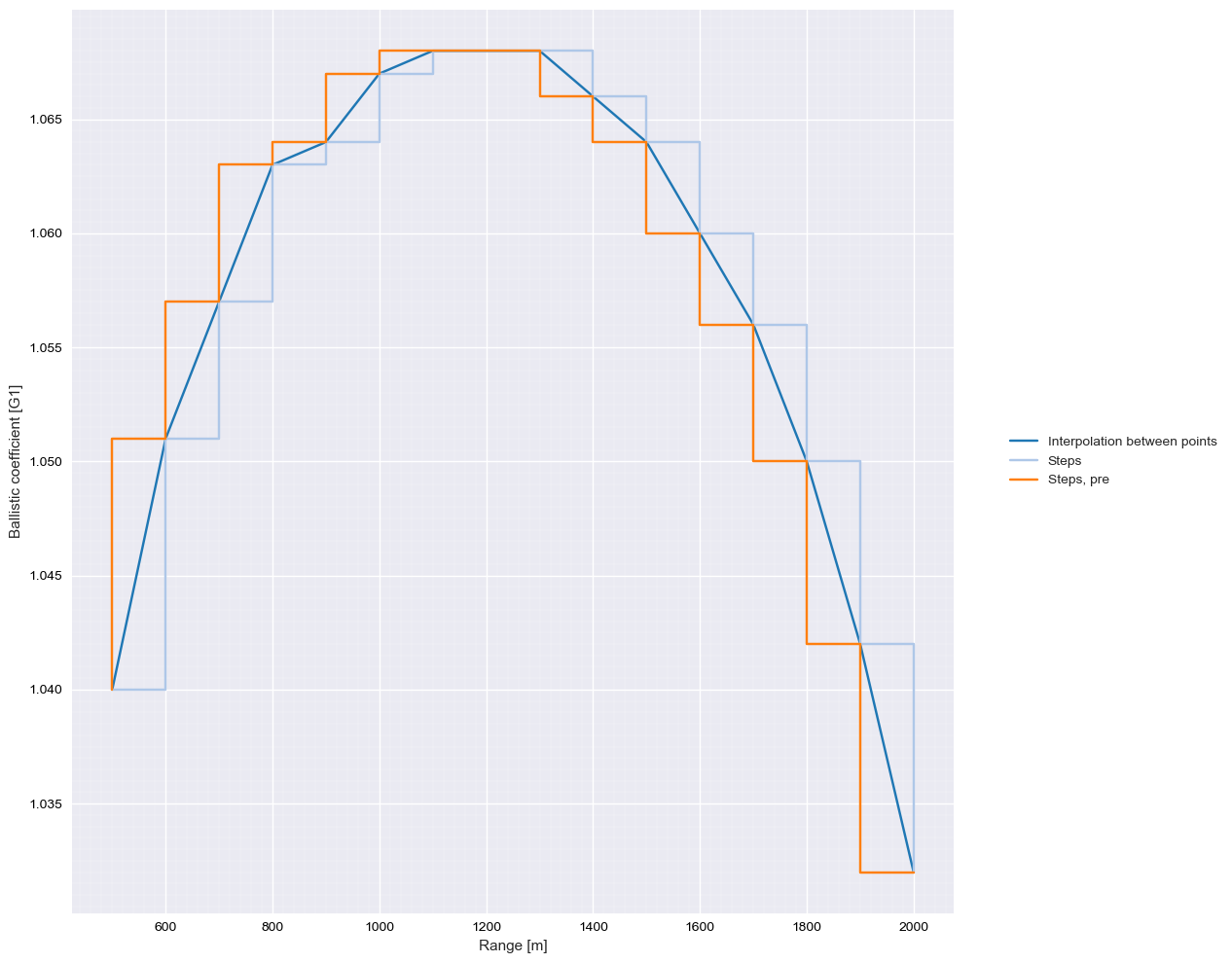
Slicing
In [21]:
BC[2:4]
Out[21]:
mesh2d
| 0 | 1 | |
|---|---|---|
| Range [m] | 700 | 800 |
| Label [unit] | 1.057 | 1.063 |
Breakpoints strictly monotone, reverse order has no effect
In [22]:
BC[::-1]
Out[22]:
mesh2d
| 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | ... | 15 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Range [m] | 500 | 600 | 700 | 800 | 900 | 1000 | 1100 | 1200 | 1300 | 1400 | 1500 | 1600 | 1700 | 1800 | ... | 2000 |
| Label [unit] | 1.04 | 1.051 | 1.057 | 1.063 | 1.064 | 1.067 | 1.068 | 1.068 | 1.068 | 1.066 | 1.064 | 1.06 | 1.056 | 1.05 | ... | 1.032 |
In [23]:
BC[6]
Out[23]:
(1100, 1.0680000000000001)
In [24]:
BC.x
Out[24]:
BreakPoints: Range [m]
| 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | ... | 15 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 500 | 600 | 700 | 800 | 900 | 1000 | 1100 | 1200 | 1300 | 1400 | 1500 | 1600 | 1700 | 1800 | ... | 2000 |
In [25]:
BC(np.arange(500, 550, 10))
Out[25]:
array([ 1.04 , 1.0411, 1.0422, 1.0433, 1.0444])
In [26]:
BC.resample(np.arange(500, 2000, 200))
Out[26]:
mesh2d
| 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | |
|---|---|---|---|---|---|---|---|---|
| Range [m] | 500 | 700 | 900 | 1100 | 1300 | 1500 | 1700 | 1900 |
| Ballistic coefficient [G1] | 1.04 | 1.057 | 1.064 | 1.068 | 1.068 | 1.064 | 1.056 | 1.042 |
In [27]:
BC.steps(BC.x)
Out[27]:
array([ 1.04 , 1.051, 1.057, 1.063, 1.064, 1.067, 1.068, 1.068,
1.068, 1.066, 1.064, 1.06 , 1.056, 1.05 , 1.042, 1.032])
In [28]:
BC.__dict__
Out[28]:
{'_d': array([ 1.04 , 1.051, 1.057, 1.063, 1.064, 1.067, 1.068, 1.068,
1.068, 1.066, 1.064, 1.06 , 1.056, 1.05 , 1.042, 1.032]),
'_options': {'extrapolate': False, 'step': False},
'_steps': x = BreakPoints(d=[ 500, 600, 700, 800, 900, 1000, 1100, 1200, 1300, 1400,
1500, 1600, 1700, 1800, 1900, 2000], label="Range", unit="m")
d = array([ 1.04 , 1.051, 1.057, 1.063, 1.064, 1.067, 1.068, 1.068,
1.068, 1.066, 1.064, 1.06 , 1.056, 1.05 , 1.042, 1.032]),
'_x': BreakPoints(d=[ 500, 600, 700, 800, 900, 1000, 1100, 1200, 1300, 1400,
1500, 1600, 1700, 1800, 1900, 2000], label="Range", unit="m"),
'label': 'Ballistic coefficient',
'unit': 'G1'}
.polyfit(): how to get a polymesh object from discrete values
In [29]:
BC.plot("+")
BC.polyfit(degree=2).plot(xlim=[500,2000])
BC.polyfit(degree=4).plot(xlim=[500,2000], ylim=[1.02, 1.08])
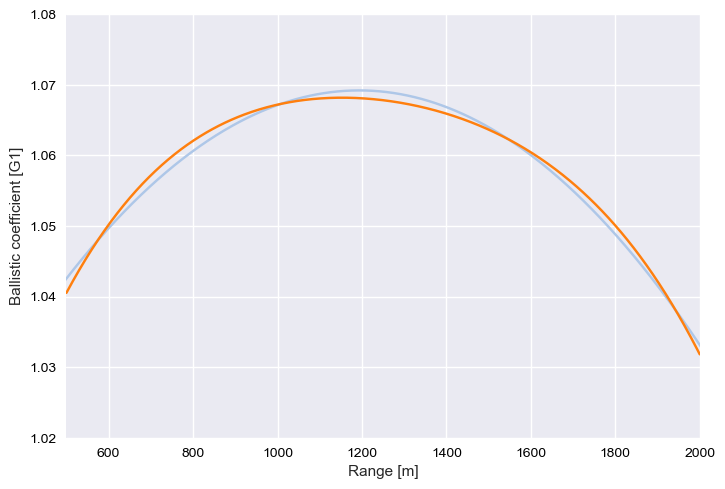
In [30]:
BC.polyfit(degree=4)
Out[30]:
In [31]:
from sklearn.metrics import r2_score
for i in range(10):
print(i, r2_score(BC.d, BC.polyfit(degree=i)(BC.x).d))
0 0.0
1 0.0698263920953
2 0.988101760851
3 0.988408030976
4 0.997727098729
5 0.998238919776
6 0.998565636158
7 0.998569159428
8 0.998610724807
9 0.998652377691
.add()
In [32]:
# prepare some data
x = [1, 2, 3, 4, 5]
y = [6, 7, 2, 4, 5]
test2 = mesh2d(x, y)
x2 = np.arange(0,60)
test = mesh2d(x2, np.sin(x2), x_label="Mon label", unit="%")
In [33]:
(test + test2)
Out[33]:
mesh2d
| 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | ... | 59 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Label [unit] | 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | ... | 59 |
| Label [%] | 5.0 | 6.84147098481 | 7.90929742683 | 2.14112000806 | 3.24319750469 | 4.04107572534 | 5.7205845018 | 7.65698659872 | 8.98935824662 | 9.41211848524 | 9.45597888911 | 10.0000097934 | 11.463427082 | 13.4201670368 | ... | 59.6367380071 |
In [34]:
test[:4]
Out[34]:
mesh2d
| 0 | 1 | 2 | 3 | |
|---|---|---|---|---|
| Mon label [unit] | 0 | 1 | 2 | 3 |
| Label [unit] | 0.0 | 0.841470984808 | 0.909297426826 | 0.14112000806 |
In [35]:
test[-3:] + test[:4]
Out[35]:
mesh2d
| 0 | 1 | 2 | 3 | 4 | 5 | 6 | |
|---|---|---|---|---|---|---|---|
| Label [unit] | 0 | 1 | 2 | 3 | 57 | 58 | 59 |
| Label [unit] | -31.2961851364 | -29.8980062588 | -29.2734719239 | -29.4849414499 | -40.90429585 | -41.115765376 | -42.2400774357 |
In [36]:
(test + test2).plot()
Out[36]:
[<matplotlib.lines.Line2D at 0x181d938240>]
In [37]:
test.steps.plot()
Out[37]:
[<matplotlib.lines.Line2D at 0x181c046630>]
In [38]:
test
Out[38]:
mesh2d
| 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | ... | 59 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mon label [unit] | 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | ... | 59 |
| Label [%] | 0.0 | 0.841470984808 | 0.909297426826 | 0.14112000806 | -0.756802495308 | -0.958924274663 | -0.279415498199 | 0.656986598719 | 0.989358246623 | 0.412118485242 | -0.544021110889 | -0.999990206551 | -0.536572918 | 0.420167036827 | ... | 0.636738007139 |
In [39]:
test.resample(np.linspace(0,60,100))
Out[39]:
mesh2d
| 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | ... | 99 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mon label [unit] | 0.0 | 0.606060606061 | 1.21212121212 | 1.81818181818 | 2.42424242424 | 3.0303030303 | 3.63636363636 | 4.24242424242 | 4.84848484848 | 5.45454545455 | 6.06060606061 | 6.66666666667 | 7.27272727273 | 7.87878787879 | ... | 60.0 |
| Label [%] | 0.0 | 0.509982415035 | 0.855858411903 | 0.896965346459 | 0.58340397644 | 0.113910235231 | -0.430285221356 | -0.805801714546 | -0.92829976264 | -0.650056648998 | -0.222663855961 | 0.344852566413 | 0.747633411784 | 0.94907077415 | ... | 0.280603366194 |
In [40]:
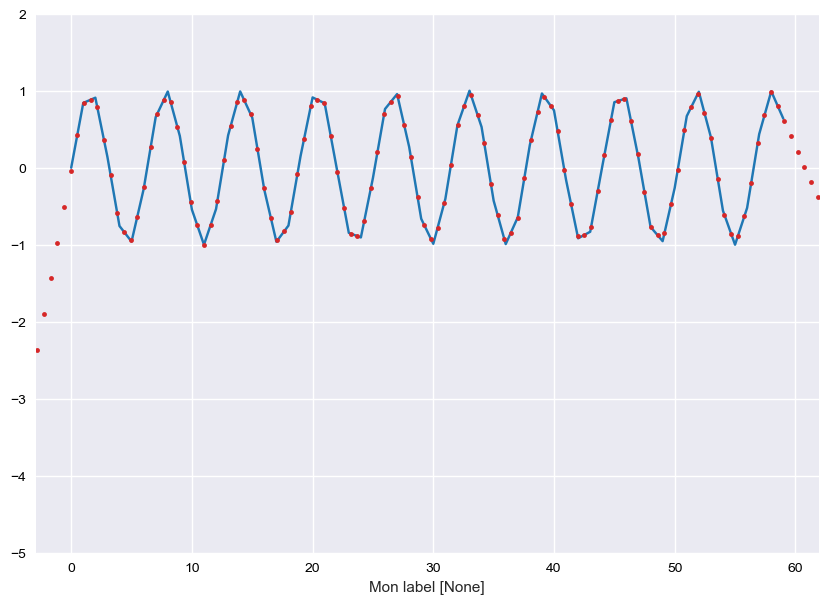
plt.figure(figsize=(12,7))
test.plot()
newX = np.linspace(-10,100,200)
test.resample(newX).plot('.', c=category20.by_key()['color'][6], lw=0.1)
plt.ylim(-5, 2)
Out[40]:
(-5, 2)
Not implemented
In [41]:
test + test2.steps
Out[41]:
mesh2d
| 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | ... | 59 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Label [unit] | 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | ... | 59 |
| Label [%] | 5.0 | 6.84147098481 | 7.90929742683 | 2.14112000806 | 3.24319750469 | 4.04107572534 | 5.7205845018 | 7.65698659872 | 8.98935824662 | 9.41211848524 | 9.45597888911 | 10.0000097934 | 11.463427082 | 13.4201670368 | ... | 59.6367380071 |
In [42]:
test.steps([-10, 2.3, 3, 3.1, 59, 58.9, 90])
Out[42]:
array([ -8.41470985, 0.6788442 , 0.14112001, 0.05132776,
0.63673801, 0.67235147, -10.40343586])
In [43]:
test.steps
Out[43]:
mesh2d
| 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | ... | 59 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mon label [unit] | 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | ... | 59 |
| Label [%] | 0.0 | 0.841470984808 | 0.909297426826 | 0.14112000806 | -0.756802495308 | -0.958924274663 | -0.279415498199 | 0.656986598719 | 0.989358246623 | 0.412118485242 | -0.544021110889 | -0.999990206551 | -0.536572918 | 0.420167036827 | ... | 0.636738007139 |
In [44]:
test.diff(n=1)
Out[44]:
mesh2d
| 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | ... | 58 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mon label [unit] | 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | ... | 58 |
| Label [unit] | 0.841470984808 | 0.0678264420178 | -0.768177418766 | -0.897922503368 | -0.202121779355 | 0.679508776464 | 0.936402096918 | 0.332371647905 | -0.577239761382 | -0.956139596131 | -0.455969095661 | 0.46341728855 | 0.956739954827 | 0.570440318868 | ... | -0.356134640945 |
In [45]:
#plt.plot(test.X, test.Y, c="b")
newX = np.arange(-10, 10, 0.001)
plt.plot(newX, test(newX), ":r")
plt.plot(newX, test.steps(newX), "-g")
Out[45]:
[<matplotlib.lines.Line2D at 0x181ba962e8>]
In [46]:
test.steps(newX)
Out[46]:
array([-8.41470985, -8.41386838, -8.41302691, ..., -0.54115269,
-0.54210883, -0.54306497])
In [47]:
import random
from numba import jit
N = 200
myMesh = mesh2d(np.arange(N)*5, [random.uniform(2.5, 10.0) for i in range(N)], x_label="MON Label")
In [48]:
@jit
def _extrapolate(self, X):
"""
"""
if X <= self.x[0]:
res = self.d[0] + (X - self.x[0]) *\
(self.d[1] - self.d[0]) / (self.x[1] - self.x[0])
elif X >= self.x[-1]:
res = self.d[-1] + (x - self.x[-1]) *\
(self.d[-1] - self.d[-2]) / (self.x[-1] - self.x[-2])
else:
res = np.interp(X, self.x, self.d)
return res
In [49]:
%timeit myMesh.extrapolate(255)
21.7 µs ± 915 ns per loop (mean ± std. dev. of 7 runs, 10000 loops each)
In [50]:
%timeit _extrapolate(myMesh, 255)
The slowest run took 5.50 times longer than the fastest. This could mean that an intermediate result is being cached.
94.3 µs ± 87.9 µs per loop (mean ± std. dev. of 7 runs, 1 loop each)
In [51]:
np.diff(test2.d) / np.diff(test2.x)
Out[51]:
BreakPoints: Label [unit]
| 0 | 1 | 2 | 3 |
|---|---|---|---|
| 1.0 | -5.0 | 2.0 | 1.0 |
mesh3d¶
In [52]:
m3d = mesh3d(x=np.arange(10), x_label="X", x_unit="X unit",
y=np.arange(10), y_label="Y", y_unit="Y unit",
d=np.random.random((10,10)), label="data", unit="data unit")
In [53]:
lerp.options.display.max_rows = 15
In [54]:
m3d
Out[54]:
mesh3d: data [data unit]
| Y [Y unit] | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | |||
| 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | |||
| X [X unit] | 0 | 0 | 0.434870352236 | 0.231482446855 | 0.323554590807 | 0.194513188669 | 0.548793345442 | 0.96149870994 | 0.844978297764 | 0.0228477172479 | 0.897581614385 | 0.374790493167 |
| 1 | 1 | 0.437402209826 | 0.587596826678 | 0.791935900857 | 0.797250085103 | 0.172353816711 | 0.307922572235 | 0.892314438845 | 0.0389261541275 | 0.152285593713 | 0.668586250984 | |
| 2 | 2 | 0.239566423457 | 0.117757222828 | 0.0778158520051 | 0.408587080639 | 0.575914282062 | 0.615422273431 | 0.0794269216571 | 0.241520040366 | 0.576474949269 | 0.428839200808 | |
| 3 | 3 | 0.401880455557 | 0.323199273872 | 0.457562472467 | 0.0141568154629 | 0.227149073091 | 0.465886627968 | 0.103495434375 | 0.432788853357 | 0.543142697729 | 0.624016873678 | |
| 4 | 4 | 0.997010442368 | 0.580724632779 | 0.78920553766 | 0.433053909205 | 0.899167356663 | 0.136601551671 | 0.321671088169 | 0.0719736098203 | 0.873654825756 | 0.0908232066389 | |
| 5 | 5 | 0.839283164546 | 0.0983105134967 | 0.336623680254 | 0.922124339553 | 0.000231907931159 | 0.299444518868 | 0.14549982256 | 0.941861267225 | 0.0852897382606 | 0.806634210453 | |
| 6 | 6 | 0.329042869533 | 0.733084098255 | 0.36561023492 | 0.837637339275 | 0.358071677098 | 0.819752114767 | 0.835915137057 | 0.612484846397 | 0.665516593392 | 0.917622397535 | |
| 7 | 7 | 0.321520224323 | 0.882292871723 | 0.131471934025 | 0.0638956540853 | 0.0575484791685 | 0.313598324598 | 0.932679129866 | 0.582086515852 | 0.271956817773 | 0.322686998655 | |
| 8 | 8 | 0.687338866596 | 0.342367197937 | 0.00667948763791 | 0.878436083913 | 0.103669100142 | 0.78076360356 | 0.794991264318 | 0.100103968489 | 0.498882006535 | 0.515767614048 | |
| 9 | 9 | 0.662384794299 | 0.984440654163 | 0.994469603672 | 0.611867904981 | 0.852381106735 | 0.846679230244 | 0.689709361867 | 0.136597804809 | 0.376000954499 | 0.960696980125 | |
In [55]:
m3d(4.5, 5.7)
Out[55]:
0.22891672933562421
Interpolation
In [56]:
m3d(3.5, 5.6)
Out[56]:
0.24804759269057369
Call method default : extrapolation above boundaries
In [57]:
m3d(9.1,9.4)
Out[57]:
1.2617807437078306
As far mesh2d, can be set with options attributes or method call .interpolate
In [58]:
m3d.options
Out[58]:
{'extrapolate': True}
In [59]:
m3d.interpolate(9.1,9.4)
Out[59]:
0.9606969801252196
In [60]:
m3d(x=3.5)
Out[60]:
mesh2d
| 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | |
|---|---|---|---|---|---|---|---|---|---|---|
| Y [Y unit] | 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 |
| Label [unit] | 0.699445448962 | 0.451961953325 | 0.623384005064 | 0.223605362334 | 0.563158214877 | 0.301244089819 | 0.212583261272 | 0.252381231588 | 0.708398761743 | 0.357420040158 |
Slicing
In [61]:
m3d[3:6]
Out[61]:
mesh3d: data [data unit]
| Y [Y unit] | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | |||
| 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | |||
| X [X unit] | 0 | 3 | 0.401880455557 | 0.323199273872 | 0.457562472467 | 0.0141568154629 | 0.227149073091 | 0.465886627968 | 0.103495434375 | 0.432788853357 | 0.543142697729 | 0.624016873678 |
| 1 | 4 | 0.997010442368 | 0.580724632779 | 0.78920553766 | 0.433053909205 | 0.899167356663 | 0.136601551671 | 0.321671088169 | 0.0719736098203 | 0.873654825756 | 0.0908232066389 | |
| 2 | 5 | 0.839283164546 | 0.0983105134967 | 0.336623680254 | 0.922124339553 | 0.000231907931159 | 0.299444518868 | 0.14549982256 | 0.941861267225 | 0.0852897382606 | 0.806634210453 | |
Garbage after that point
In [62]:
from scipy import misc
from scipy import ndimage
# http://www.ndt.net/article/wcndt00/papers/idn360/idn360.htm
from PIL import Image
from urllib.request import urlopen
import io
URL = 'https://www.researchgate.net/profile/Robert_Dinnebier/publication/268693474/figure/fig5/AS:268859169046548@1441112429791/Log-log-plot-of-ionic-conductivity-times-temperature-versus-frequency-measured-at.png'
with urlopen(URL) as url:
im = misc.imread(io.BytesIO(url.read()))
# face = misc.imread(f)
In [63]:
plt.imshow(im, cmap=plt.cm.gray, vmin=30, vmax=200)
plt.axis('off')
# plt.contour(im, [50, 200])
Out[63]:
(-0.5, 388.5, 259.5, -0.5)
In [64]:
sx = ndimage.sobel(im, axis=0, mode='constant')
sy = ndimage.sobel(im, axis=1, mode='constant')
sob = np.hypot(sx, sy)
In [65]:
sx = ndimage.sobel(im, axis=0, mode='constant')
[1] lerp article on Wikipedia